The Origins of Life itself
What do we learn about the origin of life from an information-based interpretation? One important conclusion is that on a simple understanding, the spontaneous initiation of life is extremely unlikely. Despite the optimistic 'back of an envelope' calculations considering the number of potential planets in the known Universe that may support the process, together with the several billion years available to try, a purely chance emergence of life remains very close to impossible. This chance emergence would have to be a self-assembly of a well organised network of interacting components which themselves were each quite complicated, spontaneously self-assembled structures (e.g. the progenotes hypothesised by Carl Woese in his famous "Universal Ancestor" paper (Woese, 1998)). Long before any proto-cellular biochemical mechanisms developed, there would have to be a collection of pre-biotic chemicals forming reaction networks that self-stabilised and acted as a store of working information (the system of mutual constraints that amounts to a self-sustaining causal structure (see autocatalytic sets). Suggestions for this include the lipid world (Segré et al. 2001) and RNA world (Gilbert 1986); respectively, examples of metabolism-first and genetics-first proposals (not yet theories) of the origin of life. Before talking of a hypothetical RNA-world etc., we need to consider the plausibility of pre-biotic chemistry: what went on in the pre-biotic "primordial soup" and how did it form.Primordial Chemistry
 Haldane’s Primordial Soup : illustration by InvaderXan, reproduced under the CC BY-SA 3.0 license. |
Primordial
soup is the nickname for the imagined chemically diverse
mixture that existed in standing liquid water on the earth, long before
life began (>3.7x10^9 years ago). The cartoon opposite celebrates
John Haldane's (English) model of this mixture , but could more fairly
have celebrated that of Alexander Oparin (Russian) who came to a
similar conclusion five years earlier in 1924 (though it was
Haldane
who first coined the nickname). That conclusion was that the conditions
on early earth were those needed to spontaneously create a wide range
of small organic molecules; most notably among these were amino acids -
the 'building blocks' for proteins. Actually, the idea that life
originated from self-organising biochemical cylces was first proposed
by Ernst Haeckel (1834-1919) who called the mixture Urschleim (primitive slime), so we are told by the grandee of information theory in biology Hubert Yockey* (2005) - more of that shortly. The seminal experiment conceived
and executed by Stanly Miller and Harold Urey demonstrated the veracity
of Haekel's, Oparin's and Haldane's ideas up to a point. The Miller-Urey experiment (1952) showed
that water, methane, ammonia and hydrogen gas (all abundant on the
early earth), along with heat and lots of sparks (representing
lightning) could produce a handful of amino acids. More recently, further
amino acids have been added to the list and, at least as significant,
the nucleotides (the bases for RNA and DNA) could be spontaneously
produced in a chemical 'soup', plausibly representative of the early
earth. This is, though, a very long way from the, so far merely speculated, life-forming biochemical cycles. |
Soup made, we have next to turn to whether these small organic molecules can assemble into the peptides (small proteins) and perhaps also nucleotides (little nucleic acid chains), and other components necessary for life (such as membranes). But why are these sort of molecules essential for life, anyway?
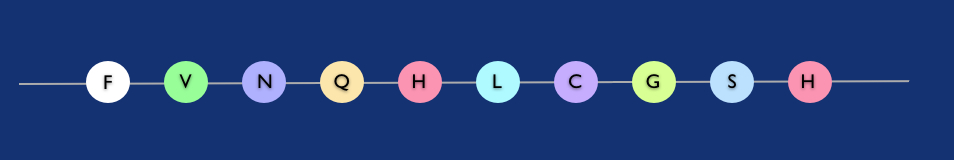
Polypeptide string: amino acids like beads (in fact the first few amino-acids of the insulin molecule).
Peptides and nucleotides in particular are so important because they are composed of strings of a small set of distinct component molecules (e.g. four in the case of RNA nucleotides). This means that a huge range of different molecules can be formed just by varying the order in which the distinct components are strung together.
Crucially, the joining of any (arbitrarily chosen) pair of amino acids or nucleotides is very nearly thermodynamically equivalent (that is any one of them can bond with any other, with the same energetic consequences). This means that the information entropy (intropy) of a polypeptide or nucleic acid molecule is decoupled from its thermodynamic entropy. Thus any sequence can be made and the probability of any sequence depends only on its length (assuming equal numbers of kinds of 'beads' are available to string together). Recall that information entropy measures the capacity of a class of forms (e.g. RNA sequences of a given length) to store information. Chains of monomers formed can either directly store information in the sequence of monomers (as in RNA) or in the shape (the form) of folded and knotted molecular structures (specifically, secondary, tertiary and, once they get really long, quaternary structure). The latter is strictly information in form, but is crucial because it is specific form that enable the special chemistry of life to occur, most especially catalysis (see physical basis of cause). On the other hand, the fact that the sequence of assembled peptides or nucleotides is thermodynamically arbitrary (equivalent) also means that there is no thermodynamic direction for what sequence will form, so particular sequences (that result in e.g. particular proteins, or RNA messages) cannot assemble spontaneously with probability greater that that predicted in the sequence paradox problem. Instead, they have to be constructed by organised chemical reactions.
This construction, rather than spontaneous assembly is the most crucial difference between living chemistry and all other chemistry: the latter is entirely guided by thermodynamic gradients, and the products are spontaneous, the former is guided by form (information) constraining chemical forces and the products are constructed. In practice, the construction requires a suite of catalysts, themselves proteins and RNAs that have to be constructed by catalysts.
Such functional molecules (the catalysts) also need to be formed at sufficient rate to get enough of them in close proximity to one another to start the precursors to biochemical reactions that establish a causal loop of self making: that is the crux-point in biogenesis.
Therefore, for life to start, catalysts have to catalyse their own making. How plausible that is depends, among other things, on how likely such molecules can form spontaneously in the first place, since at the beginning there were no catalysts to construct them - this is the original chicken and egg problem!
Back of an envelope
There are about 100 billion (10^11) stars in our galexy and we can detect about 1 billion galexies in the universe and studies of exoplanetary systems shows that 10 planets per star is a reasonable number, so roughly 10^21 planents in total are available with a few billion years to work on making life. So far we are unsure what proportion of exoplanets harbour the conditions needed to start life (not least because we do not really know the conditions required). A very strong argument can be made for two things being necessary: liquid water and relative peace (freedom from cataclysmic collisions with e.g. metiors and intense radiation), which together may narrow the field by several orders of magnitude (powers of 10). If we (hypothetically) say 1 in a thousand has the right conditions, then could it be that on many of them, life has begun? We do not yet know how fragile early life would be, so cannot calculate its chance of survival, but it is quite possible that it would need to start in many places and many times over again to build up enough self-replicating organisms that a viable population can persist. To be generous, let us hypothesise that if life is possible on a planet, then it will happen and will persist, so we can conclude there are around 10^18 chances in the whole Universe (which is really a lot).
On the other hand, even a minimal life form (as simple as possible) is likely to consist of at least several hundred different (interacting) peptides and quite likely RNA molecules (or something similar). Many of these will typically have to be of a size similar to that of the insulin molecule (50 amino acids in a row). This immediately raises the sequence paradox : to get 50 amino acids in the right order, given there are 20 kinds to choose from, is as likely as 1 chance in 10^80 trials. If there were, let us say just 100 such molecules required to start life, then by the basic laws of probability, we would need not 100 times 10^80, but 10^80 times 10^80 times 10^80.... 100 times, which makes 10^180 rolls of the dice to stand an even chance of getting all 100 molecules in place at the same time. On each planet there may be (again let's be generous) 10^12 tonnes of amino acid raw material (recalling the productivity of the Millar-Urey experiment which yielded amino acids from inorganic chemicals), that would be about 10^21 time 10^12 = 10^33 raw material molecules (using Avagadro's number and a molecular weight of about 100) to work with. This means that on each planet the 'dice can be rolled' around 10^33 / 5000 times simultaneously (each time seeing if the amino acids fell in the right order, for all 100 peptides of average length 50 amino acids). So we would need 10^180 rolls and would have about 10^30 rolls per planet times 10^18 planets = 10^48 rolls of the dice in the entire universe. But that is just in one big simultaneous rolling of dice. Suppose we could roll the dice every half a microsecond (a reasonable rate for this sort of chemical reaction), then in 4 billion years, we would get about 10^23 goes, so the total number of rolls in the whole Universe, over 4 billion years would be about 10^23 times 10^48 = 10^71. With all these trials, our chance of a single system of the right 100 peptides is 10^71 / 10^180, which is one in 10^109 and that is so small a chance that it remains inconceivable. As Paul Davies says in his book (The Origin of Life, Penguine 2003) "There is a world of difference between building blocks and an assembled structure. Just as the discovery of a pile of bricks is no guarantee that a house lies around the corner, so a collection of amino acids is a long, long way from the sort of large, specialized molecules such as proteins that life needs."
And yet, here we all are. It did happen, at least once.
* I should add, by the way, that this is my version of an argument already made very cogently by Hubert Yockey (1916-2016) and summarised in his 2005 book. Originally a radiation physicist, he turned to applying information theory to fundamental problems in biology and his work has become one of the pillars of this science. He took a very pessimistic view of our ability to reconstruct the origin of life and unfortunately his skepticism was grasped and misused by "Intelligent Design" believers, probably harming his reputation. One could interpret this theme as an attempt to carry on where he left things - a search for a scientific explanation that overcomes the great obstacles that he so clearly delineated.
Natural ways to make it more likely
The terrible odds explained above can be interpreted in two different ways: either life on earth is effectively a miracle, or the simple calculations above are misleading because somehow assembling the structure of life is made more likely by the very process of attempting it. At first, this latter seems a weird and implausible answer, but there are some good (scientific) reasons for believing in it. One of them is that if life really is an information process, then its self-assembly could have followed a recursive proceedure of bootstrapping. In language that is now familiar, if life is computation, then perhaps it started by 'booting up'. Of course, before there was life, nothing could be 'attempting' anything; it would all have to rely on spontaneous (thermodynamically controlled) action, so what this idea really means is that each step on the way makes the next step more likely than the conditional probability would suggest.
To briefly illustrate the idea, consider manufacturing for a moment. A set of machines make parts for e.g. an engine. But where did those machines come from? They were made by other machines which fabricated the components needed for the machines we started with. The machine-component making machines were made by other machines ... and so on. Carpentry illustrates the bootstrapping behind this: One of the first things a keen carpenter will make is a sliding table saw sledge. This handy device enables them to accurately cut forms that assemble into a wide range of jigs - structures that function as tools in the construction of other structures. One of the first uses of the first jigs the carpenter makes, is the construction of more sophisticated jigs. Eventually, with a suitable array of jigs hanging on the workshop wall, the carpenter is limited only by their imagination (and wood supply). We can imagine that life emerged as relatively simple molecules were somehow useful in the making of more complicated (information rich) molecules which formed supramolecular structures that eventually constituted the first life. Of course there was no carpenter (or even a blind watchmaker). As explained on the bootstrapping page, all the engineering examples cheat by 'borrowing' information from a separate source or store. That cheat was not available to the origin of life.
But there is a cheat that we can permit. It is easily understood from the carpentry illustration: the only thing that mattered about those jigs was their physical form, and only the functional parts of that form mattered. For example, the saw sledge could have lots of holes in it, but it would still work fine. So it is with catalytic molecules: only certain shapes within the molecule perform the catalytic function; the rest of it is termed 'scafold', meaning it is just the structure around which the functional bits form and the details of its structure don't really matter. This is very helpful becaue it means we don't need to find a particular sequence of monomers for the whole molecule, only for the functional parts of it and the rest has a more relaxed specification: several alternative sequences will do for the scaffold.
One of the 'holy grail's of the RNA world hypothesis is a ribozyme, specifically an RNA molecule that catalyses its own formation. Such a molecule can be synthesised in the lab, but nothing yet observed in the natural world comes close. However, its function is dependent on a small set of patterns - simple forms such as rings and hairpins (stem-loop patterns). RNA is like sticky string - you don't need much before it is sticking to itself and making a tight little ball. But not quite because the sticking is actually the formation of hydrogen bonds which only works between particular pairs of bases (we will now refer to nucleotides by their unique bases). G-C, A-U (RNA only), A-T (DNA only) and T-G (DNA only) are all Watson-Crick base pairings (the same that join the two DNA strands into a double helix) and G-U (RNA only) is the more subtle 'wobble base pairing', Base pairing is an information constrained relationship between patterns (again see information bootrapping) in which a base is held within 2.5 Å of its pairing partner further along the chain (for comparison, a DNA double helix is 10 Å across). Information constraint works in two respects: the specific placement of nitrogen and oxygen atoms on the purine and pyrimidine bases (their form) lets them join by sharing hydrogen atoms, but only the pairs listed have the configuration to do this, so base pairing is very selective.
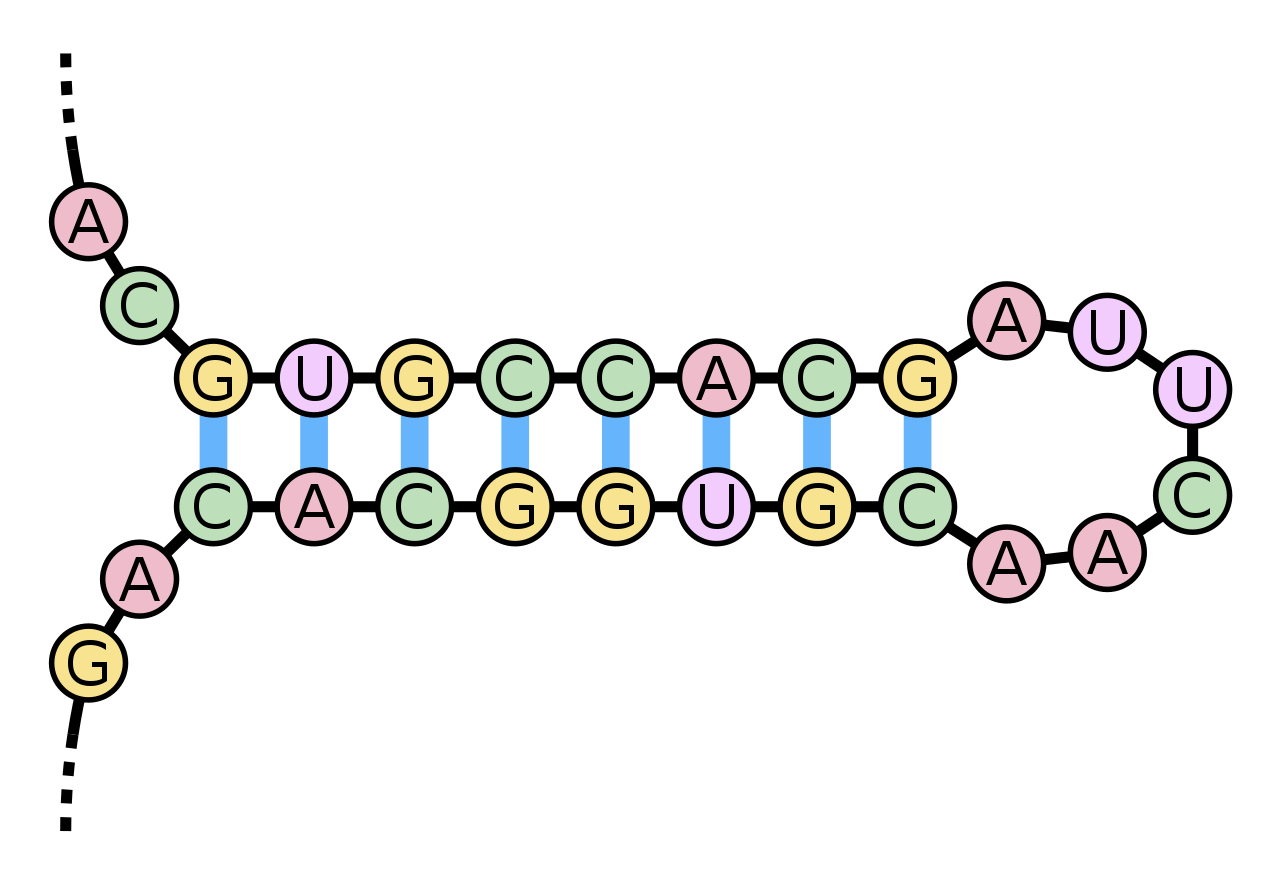
A stem and loop (hairpin) taken from Wikipedia (the blue bands are hydrogen bonds).
The A opposite A cannot pair and the resulting separation forms the UUCA sequence into a loop.
The A opposite A cannot pair and the resulting separation forms the UUCA sequence into a loop.
Sequences that include lengths that can form these pairings, intersperseded with those that cannot, often produce hairpins (as above) and these patterns are typically functional, often having catalytic effect, able to cleave and contribute to joining (ligation) and they are quite common.
Once there are lots of hairpins floating about (typical length 20 bases), the probability that some will join in small groups is reasonably high. Is it a big leap to join three of these together and, with a bit of rearrangement of a few bases, get e.g. a tRNA molecule? One of the virtues of the loop is that bases on its outer edge (ACUU in the diagram above) are unconstrained, so could provide a signal or message in the form of a pattern to match (e.g. by weak binding with another molecule). This way little loops can act as the variable shape of a door key - specific to a particular lock. That is the role of the anti-codon triplet in one of the three loops of tRNA. This talk of tRNA is premature and quite misleading, though. First, whilst rival theories for the origin of tRNA have yet to be resolved (see e.g. Di Giulio, 2020), neither involve merely joining hairpins and the way tRNA is fabricated in extant cells is really quite complicated, secondly tRNA would have no function at the stage of RNA-world and indeed as a general point, in RNA world, the sequence of bases serves no coding purpose at all, but rather is entirely a matter of creating the right secondary and tertiary structure (molecular form) to confer enzymatic functions.
We should instead look for other small non-coding RNA molecules with function and the top candidate (presently) is the hammerhead ribozyme (see de la Peña et al, 2017). To see if catalytically functional RNAs could arise naturally, Wilson and Szostak (1999) used a modification of the PCR reaction method to selectively amplify random strands of RNA with 220 bases (10^15 of them). With very strong selection (and help from a DNA template) about 100 of the original population were effective RNA ligase catalysts (they could join bits of RNA together). Further 'natural' selection among these resulted in some ligase RNA molecules that were as effective as modern protein equivalents. It should be noted that although 10^15 variations seems a lot, the number of possible variations among 220 base RNA sequences is 10^132, so this success was found with a tiny subset of possibilities. On the other hand, 220 base sequences, though short in terms of those prevalent in living systems are really long for spontaneous abiotic assembly. Hairpins are typically around 20 bases long and hammerheads no more than 40.
So, how likely is a soup of diverse ribozymes and how likely is this to result in an autocatalytic set? Further, how likely is that to survive multiple generations of reproduction, given the propensity for components to 'mutate' via errors in mutual construction (for example ligating similar but the wrong component micro RNAs)? We can only answer these questions with probability theory applied to quantifications of information transfer.
A few thing seem certain. The molecules must be enclosed by a selectively permeable membrane to have any chance of meeting one another often enough for biochemistry to develop (see the lipid membrane on the autopoiesis page). One of the RNAs that must be created is a reasonably efficient RNA polymerase ribozyme. Quite likely cleaving ribozymes would be needed too. There is a tendency in RNA world theory circles to neglect the possible role of polypeptides and other biomolecules that may be present and act to help the RNA-based autopoietic system to develop: they might have a role. Of most significance would be some way of deriving work out of energy from the environment to power some of the reactions, but these matters are highly speculative at present.
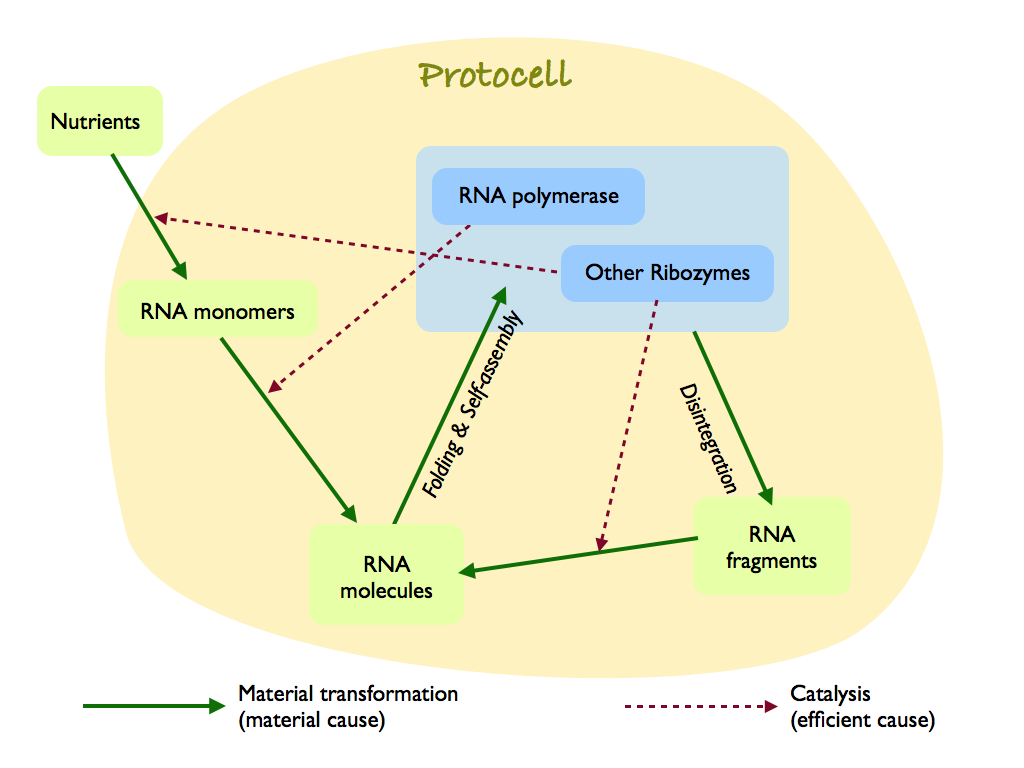
A hypothetical protocell, which is a membrane encapsulated RNA world ribozyme network. I have drawn it as, what I now call, a Hofmeyr diagram (after Hofmeyr, 2017). It is a reinterpretation of Kawamura's RNA-based life-like system (Kawamura, 2019), but adds explicit separation of material and efficient cause and the action of ribozomes other than the central RNA polymerase, which help build RNA monomers and repair RNA molecules that inevitably disintegrate. Kawamura recognised that the chemistry needed a enclosure to keep the concentration of interacting molecules high (this is made explicit by Luisi's autopoietic proto-life systems (see the autopoiesis page for both Luisi and Hofmeyr references).
Hieronymous and Müller, 2021
I am actively working on the text here (11-05-21)
Thinking of early life only in terms of information transfer translation (genes to proteins) and inheritance via the central dogma is where we run up against Eigen’s paradox: no efficient enzymes are possible without accurate information templates (e.g. nucleic acids), but no accurate information templates are possible without efficient enzymes (for error correction): more chicken and egg, this time attributed to Manfred Eigen (1927-2019). It is based on the fact that reproduction involves errors - mutations that corrupt the information needed to make the enzymes that prevent the corruption of the information, and so on ad infinitum. This is a conundrum that cannot be solved, at least not directly, it must be negotiated around.
Stuart Kauffman provides a good introduction to efforts made to understand the origin of life (here).
Many of the proposed mechanisms seem to fail in practice, becasue of biochemical difficulties and in particular an apparent bottle-neck in trasmitting the information necessary to construct a complex chemical system, reliably and repeatedly.
Understanding what life is, in the most fundamental sense possible, is a good starting point for understanding how it arose originally. There are several, as yet unconfirmed, theories about the origin of life on earth, but what about more generally: how can it emerge independently of the earth? To answer this question, we must first establish what specifically and sufficiently constitutes life.
Life is information processing
The first point is that all the life we know of consists of a complex network of highly coordinated chemical reactions. If all the right chemicals are available, but they are not reacting, or not reacting in the right place at the right time, then, no life will be found. If there are not the necessary chemicals, the same will be true. The difference between the living and the dead is that the living constitute a particular set of reactions, organised into a particular configuration in space and time. A dead organism is also a network of chemical reactions, but these are different and less coordinated: they are the reactions of decay and chemical return to equilibrium with their surroundings. Indeed one defining feature of life is that it is out of chemical (and therefore energy) equilibrium with its environment, but then so is a star. There is no complex organisation of chemical reactions in a star (as we understand them) and their structure is maintained as an average of a very large number of random configurations and interactions among their component parts. Life is characterised by a surprising lack of randomness in configuration: it is a persistent pattern, which is the embodiment of information (formative information). At the other end of the scale, we could point to a crystal, which is highly organised (an ordered array of atoms), but is also static, not dynamic with reactions, and it is so well ordered as to be remarkably simple and therefore embodying remarkably little information. Somewhere between the ferocious reactivity of a star and the ordered timelessness of a crystal lattice, lies something very special: a balance between dynamism and retention of complex pattern. Here we find life.
Although it has been notoriously difficult to pin down precisely what it is that makes life so distinctive and remarkable, there is general agreement that it has to do with information (indeed Manfred Eigen is not the only scientist to say that "life is information"). The unique informational narrative of living systems suggests that life may be characterised by context-dependent causal influences, and, in particular, that top-down (or downward) causation—where higher levels influence and constrain the dynamics of lower levels in organisational hierarchies—may be a major contributor to the hierarchal structure of living systems. Here, we propose that the emergence of life may correspond to a physical transition associated with a shift in the causal structure, where information gains direct and context-dependent causal efficacy over the matter in which it is instantiated. Such a transition may be akin to more traditional physical transitions (e.g. thermodynamic phase transitions), with the crucial distinction that determining which phase (non-life or life) a given system is in, requires dynamical information and therefore can only be inferred by identifying causal architecture. We discuss some novel research directions based on this hypothesis, including potential measures of such a transition that may be amenable to laboratory study, and how the proposed mechanism corresponds to the onset of the unique mode of (algorithmic) information processing characteristic of living systems. One thing is certain, though: life necessarily continually maintains and renews its informaiton-structure. This process, observed as self-repair and reproduction is termed autopoiesis: a unifying concept that is described here. Life would not exist without this capability.
Here is a fun description featured in the popular press (and YouTube).
Also, we like this website: Exploring Life's Origins - it's a great graphical explanations of pre-biotic & RNA world theories. Here is a good introductory article in The Scientist Magazine that is still rather up-to-date. We Have more pages in preparation for this subject area; it is just taking a bit of time! For now, we recommend the recent works of Christoph Adami and also Sara Walker and colleagues.
References
Camprubí. E., de Leeuw, J.W., C.H. House, F. Raulin, M.J. Russell, A. Spang , M.R. Tirumalai, F. Westall. (2019). The emergence of life. Space Sci. Rev. 215:56. https://doi.org/10.1007/s11214-019-0624-8
Di Giulio, M. (2020) An RNA ring was not the proginator of the tRNA molecule. J. Mol. Evol. 88:228-233.
de la Peña, M., García-Robles, I. and Cervera, A. (2017). The Hammerhead Ribozyme: A long history for a short RNA. Molecules. 22, 78. doi: 10.3390/molecules22010078 .
Gilbert, W. (1986). Origin of life: the RNA world. Nature, 319; 618.
Hieronymous, R. and Müller, S. (2021). Towards higher complexity in the RNA World: Hairpin ribozyme supported RNA recombination. ChemSystemsChem. e2100003.
Hofmeyr, J.H.S., 2017. Handbook of Anticipation: Theoretical and Applied Aspects of the use of Future in Decision Making. Springer. chapter Basic Biological Anticipation. 11, pp. 219–233.
Kawamura, K. (2019). A non-paradoxical pathway for the chemical evolution toward the most primitive RNA-based life-like system. in Evolution, Origin of Life, Concepts and Methods. Springer.
Segre, D., Ben-Eli, D., Deamer, D.W., Lancet, D. (2001). The lipid world. Orig. Life
Evol. Biosph. 31, 119–145.
Wilson, D.S. and Szostak, J.W. (1999). In vitro selection of functional nucleic acids. Ann. Rev. Biochem. 68; 611-647.
Woese, C. (1998). The universal ancestor. Proc. Nat. Acad. Sci. USA. 9;95(12):6854-9.
Yockey, H. P. (2005). Information Theory, Ecolution and the Origin of Life. Cambridge University Press.
This Theme aims to:
- Develop an understanding of the origin of life (in general) using an information-based approach;
- Describe the development of life in terms of information and
complexity.